Many of you may remember, a while ago I had written about my experiences with AWS. Recently, I was asked to launch a Google Cloud Instance,perform some tasks, output the files to a Google cloud bucket, and then shut down the instance. It was a fun experience as I had never used Google cloud and was curious to use it, and compare and contrast it with AWS. The specs were:
- Load an Ubuntu OS.
- Create the instance in the Western US region.
- Specify a 10 GB boot disk.
- Explicitly specify option to delete boot disk when the VM instance is deleted.
- Provide read-write access to Google Cloud Storage.
- Run a startup script on the instance as soon as it is launched that does the following:
- – download some publicly available tools and install them
- – run a compute job and copy the output to google cloud bucket and share it publicly
- – Get generic memory/availability info about the instance, output it to a file and upload it to the cloud bucket.
- – shutdown the instance
Google cloud services are very similar to the AWS and if you are familiar with AWS, Google Cloud Platform for AWS Professionals can be a really useful site. While the syntax and semantics of the SDK, APIs, or command-line tools provided by AWS and Google Cloud Platform, the underlying infrastructure and logic is very similar.
One of the first things you need to do is download, install and initialize the cloud SDK on your local machine. It contains gcloud, gsutil, and bq, which you can use to access Google Compute Engine, Google Cloud Storage, Google BigQuery, and other products and services from the command-line. You can run these tools interactively or in your automated scripts (as you will see later in this example). The SDK has a set of properties that govern the behavior of the gcloud command-line tool and other SDK tools. You can use these properties to control the behavior of gcloud commands across invocations.
You also need to create a gcloud account if you dont have one – and its really helpful if you have a gmail account that you can connect it to. This way when you initialize gcloud on your local machine, you can authorize access to Google Cloud Platform using a service account. The gcloud console is very similar to the AWS dashboard and you can view/control all your instances/resources from the console. The console is an easy to use interface and quite intuitive. Most of what you can do from the console can be done from the command line or using a Rest API. However, one of the things you cannot do from the command line is create a ‘Project‘ – and this is something that is different in gcloud compared to AWS. You must first create a project, get a project id which you have to reference when you spin up instances directly from the command line.
Initializing/configuring the SDK is fairly straightforward- just follow the instructions. I set up my default region to be us-west1a and set up my default project as “stanford”. I was given a project id “stanford-147200″ and I needed to reference that every time I wanted to access an instance associated with that project, start a new instance associated with that project etc. Note: you can set up multiple configurations for multiple projects and move between them quite easily because of this partitioning into “projects”. Its like creating multiple virtual environments!
If you want to store the output from the jobs run on the instance, you need to set up a cloud bucket. This is analogous to to s3 on AWS. This can be done via the console or the command line using the gsutil. You can create a bucket directly from the instance as well (as part of a script) -which is what I will do. Note: when you launch an instance, you have to specify the project id, and the bucket is created for that project.
So lets get started. I have SDK and gsutil installed on my machine. My project id is “stanford-147200″. I can launch an instance in the Western zone with an Ubuntu OS and specific properties with the following command:
gcloud compute --project "stanford-147200" instances create "stanford2" \ --zone "us-west1-a" \ --boot-disk-auto-delete \ --machine-type "f1-micro" \ --metadata-from-file startup-script="startup.sh" \ --subnet "default" --maintenance-policy "MIGRATE" \ --scopes default="https://www.googleapis.com/auth/cloud-platform" \ --tags "http-server","https-server" \ --image "/ubuntu-os-cloud/ubuntu-1404-trusty-v20161020" \ --boot-disk-size "10" --boot-disk-type "pd-standard" \ --boot-disk-device-name "stanford2"
So what do the options mean? – breaking it down:
gcloud compute : using the command line interface to interact with the Google compute facility
–project “stanford-147200” instances create “stanford2” : create an instance called ‘stanford2’ under project id “stanford-147200”
–zone “us-west1-a” :specifically create the instance in the Western US Region
–boot-disk-auto-delete : Delete the boot disk when VM instance is deleted -note this is the default
–machine-type “f1-micro” : specify machine type from the choice of publicly available images
–metadata-from-file startup-script=”startup.sh” : metadata to be made available to the guest operating system running on the instance is uploaded from a local file
–subnet “default” : Specifies the network that the instances will be part of. Default is “default”
–maintenance-policy “MIGRATE” :Specifies the behavior of the instances when their host machines undergo maintenance. The default is MIGRATE.
–scopes default=”https://www.googleapis.com/auth/cloud-platform” : specifies the access scopes of the instance- I will allow it to access all cloud API’s- this will allow it to read/write from the Google storage/BigQuery and all other Cloud services. This is very wide access – One can specify it to only access specific services
–tags “http-server”,”https-server” : allow http and https traffic ie open port 80 and 443.
–image “/ubuntu-os-cloud/ubuntu-1404-trusty-v20161020” : Use this publicly available image to create your instance -Using image with Ubuntu OS
–boot-disk-size “10” –boot-disk-type “pd-standard” : specify boot disk size to 10GB
–boot-disk-device-name “stanford2” : The name the guest operating system will see for the boot disk as.
You can go to the cloud console and see your instance being booted up, and its CPU usage. Furthermore, the instance will upload the run the startup script we have specified, change its permissions and run it. So what is in the Startup-script? Its a bash script :
#! /bin/bash # Description: SCGPM Cloud Technical challenge (2/2). # This is the startup script that is called by the Google # Compute Instance launcher script. # Author: Priya Desai # Date: 10/25/2016 ## behave like superuser sudo apt-get update ## usually need to make sure the the java version installed is 1.8 or higher. Download the JDK from Oracle website # thanks to this StackOverflow post How to automate download and installation of Java JDK on Linux?. sudo wget --no-check-certificate --no-cookies --header "Cookie: oraclelicense=accept-securebackup-cookie" -qO- http://download.oracle.com/otn-pub/java/jdk/8u5-b13/jdk-8u5-linux-x64.tar.gz | tar xvz # create symbolic link to allow update of Java/access the directory without changing the rest sudo ln -s jdk1.8.0_05 jdk #cd ~ ## Install picard-tools sudo wget https://github.com/broadinstitute/picard/releases/download/2.7.0/picard.jar -O picard.jar ## Copy bam & bai files from public bucket sudo gsutil cp gs://gbsc-gcp-project-cba-public/Challenge/noBarcode.bai . sudo gsutil cp gs://gbsc-gcp-project-cba-public/Challenge/noBarcode.bam . ## Run picard-tools BamIndexStats and write results to "bamIndexStats.txt" sudo jdk/bin/java -jar picard.jar BamIndexStats INPUT=noBarcode.bam >> bamIndexStat.txt #whoami>>files.txt echo "Memory information": >> instance_info.txt ## Append memory info for current instance to "instance_info.txt" free -m >>instance_info.txt echo " " >> instance_info.txt echo "cat /proc/meminfo:">> instance_info.txt cat /proc/meminfo >>instance_info.txt echo " " >> instance_info.txt echo " " >> instance_info.txt echo "Filesystem information": >> instance_info.txt ## Append filesystem info for current instance to "instance_info.txt" df -T>>instance_info.txt echo " ">>instance_info.txt mount >>instance_info.txt echo "mount output: " ## Upload the two output files to Google Cloud Storage bucket ##This bucket was created earlier- Can create a bucket in the script using gsutil mb sudo gsutil mb gs://stanford-cloud-challenge-priya/ sudo gsutil cp *.txt gs://stanford-cloud-challenge-priya ## Shutdown instance. Note: this just shuts down the instance-not delete it. sudo shutdown -h now
Most of the script was reasonably straight forward, but its important to note the following interesting points:
- how to install Java 1.8 (needed to run Picard tools) on the command line: Oracle has put a prevention cookie on the download link to force you to agree to the terms even though the license agreement to use Java clearly states that merely by using Java you ‘agree’ to the license….but there is a workaround -thanks to the above StackOverflow site! I could get Java 1.8 to install be using: “sudo wget –no-check-certificate –no-cookies –header “Cookie: oraclelicense=accept-securebackup-cookie” -qO- http://download.oracle.com/otn-pub/java/jdk/8u5-b13/jdk-8u5-linux-x64.tar.gz | tar xvz”
- The cloud storage bucket can be created in the script, but in order to make it publicly accessible, you need to set the permissions correctly. You can do this on the console or on the command line using gsutil. I do this in the script itself. My bucket is called ‘stanford-challenge-priya’ , and to make the bucket is accessible as: gs://stanford-challenge-priya/ , I edit the permissions with:
gsutil acl ch -u AllUsers:R gs://stanford-cloud-challenge-priya/
3 . Its important to realize that the user_script runs on the instance as “root”- You can see this by getting the output of whoami. However, when you log into your instance, you log in as a user (with your username). Hence you will not see the files that your script may have downloaded- but if you change to superuser status, you can see the files that have been downloaded.
4. You can shutdown the instance, after your jobs have completed using:
sudo shutdown -h now
Note- this only shuts down the instance , it does not delete it. So you could restart it and you will find all your files still there.
5. user_script is a great way for you to remotely, launch an instance, configure it, run jobs and shut it down so you don’t waste money keeping an instance running. If you have very complex environments that take a while to configure, you could create an instance with the configuration you like and save it as an image, give it a name and then launch that image as you need, saving you trouble of having to configure it every time!
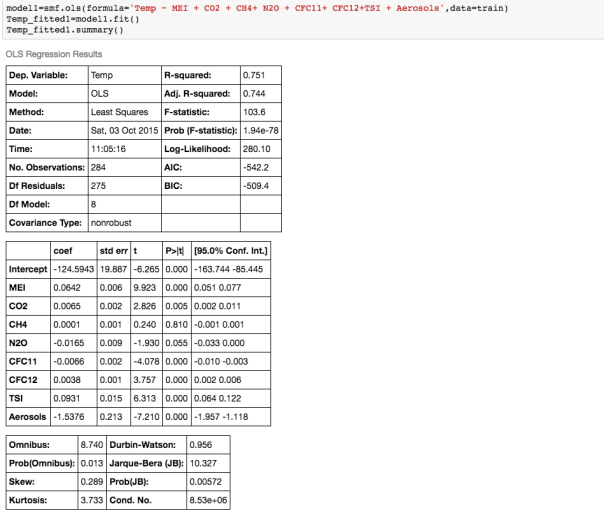
 We can see that there is a really high correlation between N2O and CH4: corr(N2O,CH4)=0.89
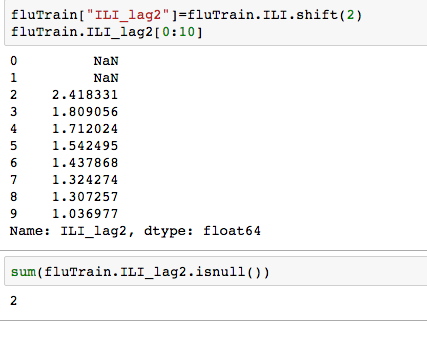
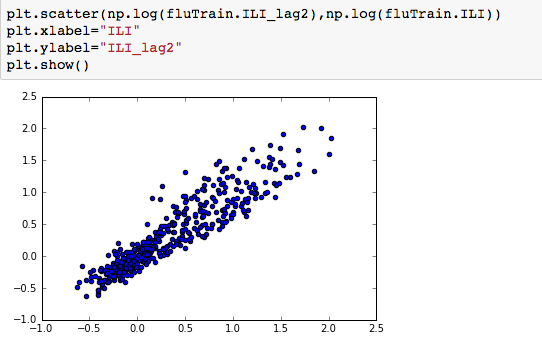
We can see that there is a really high correlation between N2O and CH4: corr(N2O,CH4)=0.89 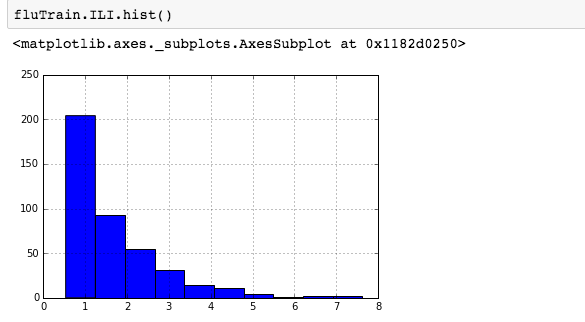 suggesting the distribution is right skewed- ie most of the ILI values are small with a relatively large number of values being large. However a plot of the natural log of ILI vs Queries shows that their probably is a positive linear relation between ln(ILI) and Queries.
suggesting the distribution is right skewed- ie most of the ILI values are small with a relatively large number of values being large. However a plot of the natural log of ILI vs Queries shows that their probably is a positive linear relation between ln(ILI) and Queries.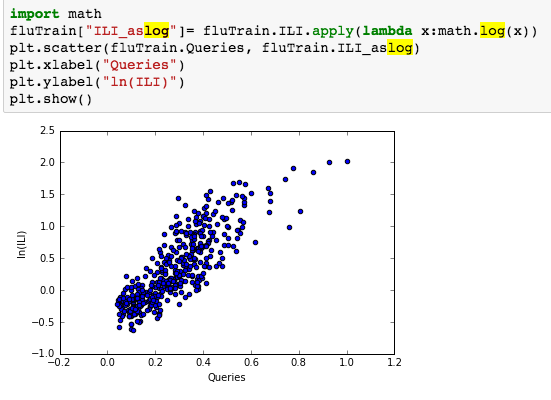


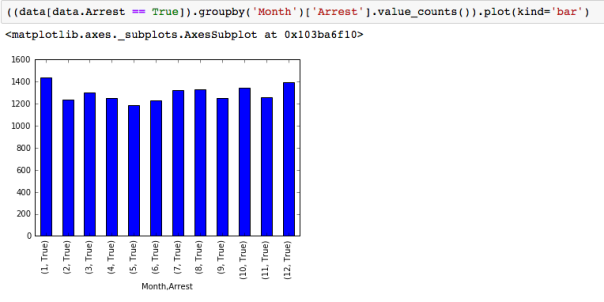
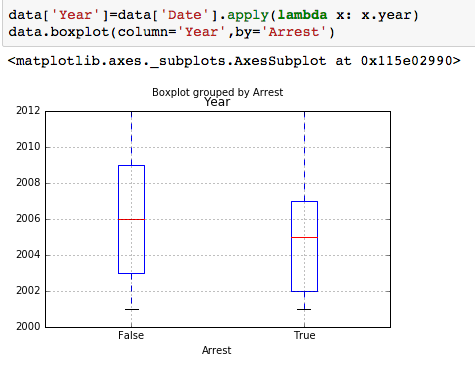
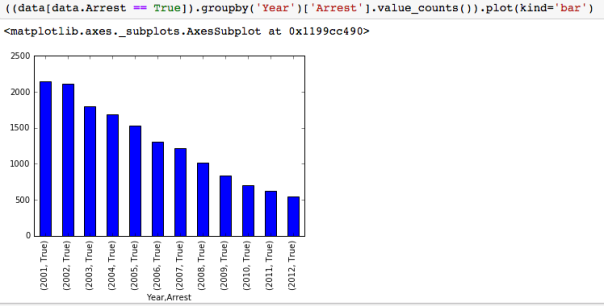
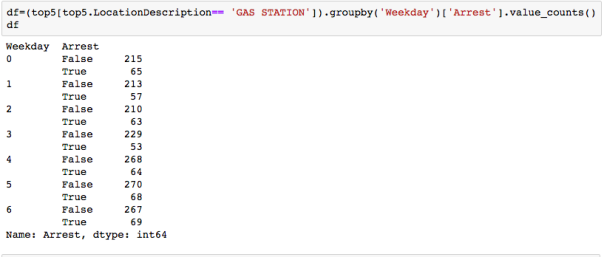

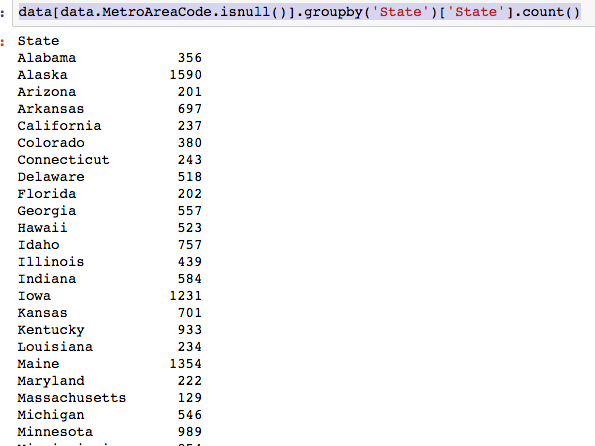


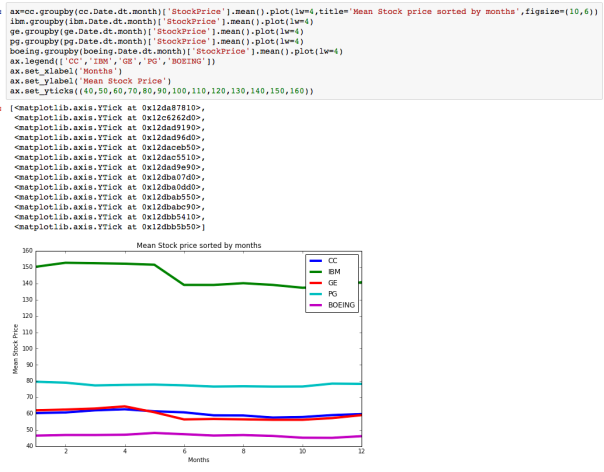
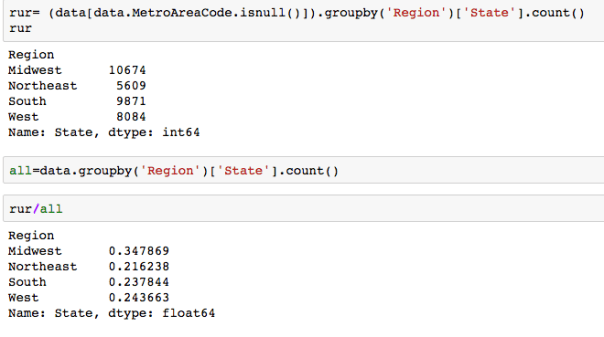 The dataset with stock prices ( see Stock_dynamics.ipynb) is really useful the play around with different plotting routines. Visualizing the stock prices of the five companies over a 10 year span -and seeing what happened after the Oct 1997 crash:
The dataset with stock prices ( see Stock_dynamics.ipynb) is really useful the play around with different plotting routines. Visualizing the stock prices of the five companies over a 10 year span -and seeing what happened after the Oct 1997 crash: Using groupby to plot monthly trends:
Using groupby to plot monthly trends: